TR&D 2 (annotation) aims to accelerate biomodeling through enhanced annotation of models, simulation experiments, and simulation results. As models accumulate in public repositories, there is an opportunity to reuse models for new studies and to combine models into comprehensive meta-models of entire biological systems. However, it is currently challenging to reuse and combine models because few models are reproducible or understandable. Consequently, modelers currently waste huge amounts of time trying to understand, reproduce, and combine models published by other modelers, including other modelers in the same research group. To make it easier to understand, reproduce, and combine models, we must make the assumptions, meanings, and limitations of models explicit.
We have begun work toward these goals by delivering a set of standards and technologies around semantic annotation. First, we have collaboratively developed the OMEX Metadata Standard, so that best practices and expectations for how to annotate models are established and agreed on by the modeling community.
Next, we have delivered a set of software tools that use this standard to enable users to appropriately annotate models, and then use these annotations to retrieve appropriate models. Details are provided below under software.
To ensure these tools accelerate biomodeling, this TR&D includes a number of Collaborative Projects which need enhanced annotation schemas and tools to understand, reproduce, reuse, and merge their models. These Collaborative Projects will push us to develop user-friendly graphical interfaces to our tools, and we will pull the Collaborative Projects to use our new annotation standards and software.
We have also established a critical Service project that provides curation capabilities. Working with PloS Computational Biology on Improving reproducibility in computational biology research, we have established a pipeline whereby authors submitting papers with models, can, as part of the peer review process, validate whether or not their model is reproducible. We produce a reproducibility report, and this can then be used to promote and better share reproducible models in publications.
Moving forward, we will continue to expand our curation work, and develop additional annotations for model credibility. In addition, we are exploring new ways to merge and connect models across different modeling paradigms. The methods, tools, and services provided by this TR&D will help modelers discover models for new studies, better understand published models, and augment and merge models to test new hypotheses about physiology and pathophysiology.
Software
libOMEXMeta C++ and Python libraries
With the growing complexity and quantity of biosimulation models, annotation tools that can accommodate a variety of modeling languages and simulation environments were needed to encourage models which could be easily understood and uniquely [...]
NLIMED
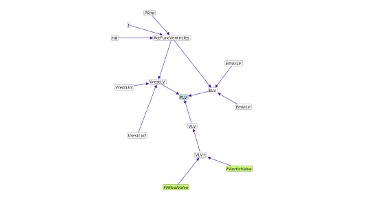
The Natural Language Interface for Model Entity Discovery in Biosimulation Model Repositories(NLIMED) was developed by Yuda Munarko to convert natural language queries into the SPARQL syntax which is typically used to search semantic annotations [...]
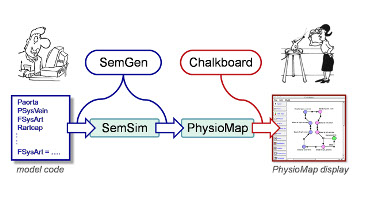
SemGen
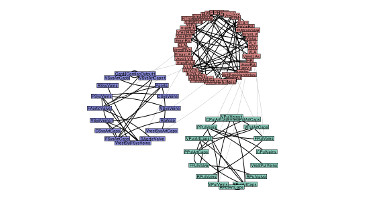
SemGen is an experimental software tool for automating the modular composition and decomposition of biosimulation models. SemGen facilitates the construction of complex, integrated models, and the swift extraction of reusable sub-models from larger ones. [...]
Standards
Formalized OMEX metadata specification
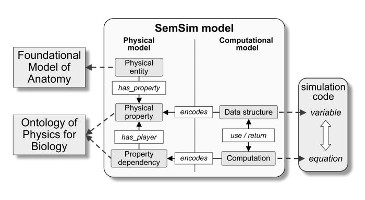
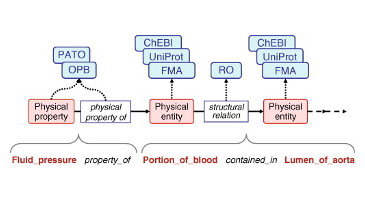
As a common language to represent annotations for models, we have developed, in close collaboration with the COMBINE community, the OMEX metadata specification. This standard provides not only the specification of sharable, RDF formatted [...]
Enhanced SED-ML libraries and specification
The Simulation Experiment Description Markup Language (SED-ML) is a standardized, community-based format that explicitly describes simulation processes for biochemical models, ensuring that researchers who are interested in a published model can efficiently reproduce and [...]
Collaborative projects
Multiscale mechanistic simulation of acetominophen-induced liver toxicity
James Sluka The Biocomplexity Institute Indiana University Bloomington, IN, USA
Alzheimer modeling
Jean-Marie Bouteiller Assistant Professor Department of Biomedical Engineering University of Southern California Los Angeles, CA, USA
BioModels model repository
Henning Hermjakob The European Bioinformatics Institute Cambridge, UK
CompuCell3d multiscale modeling
Drs. Maciek Swat and James Glazier The Biocomplexity Institute Indiana University Bloomington, IN, USA
PRiME: Program for Research on Immune Modeling and Experimentation
Stuart Sealfon Professor Neurology, Neurobiology, and Pharmacology & Systems Therapeutics Icahn School of Medicine at Mount Sinai New York, NY, USA
Simtk
Dr. Joy Ku Stanford University Stanford, CA, USA
Systems Biology Center New York
Ravi Iyengar Professor, Department of Pharmacology and Systems Therapeutics Icahn School of Medicine at Mount Sinai New York, NY, USA
Virtual Cell modeling environment
Leslie Loew Professor University of Connecticut Health Center Farmington, CT, USA
Service projects
PRiME: Program for Research on Immune Modeling and Experimentation
Stuart Sealfon Professor Neurology, Neurobiology, and Pharmacology & Systems Therapeutics Icahn School of Medicine at Mount Sinai New York, NY, USA
Team