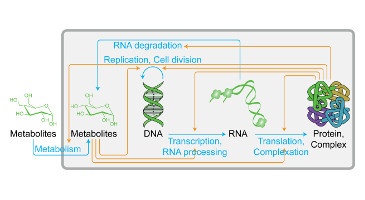
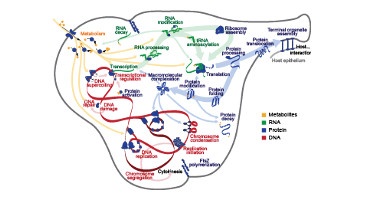
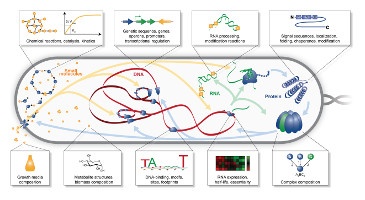
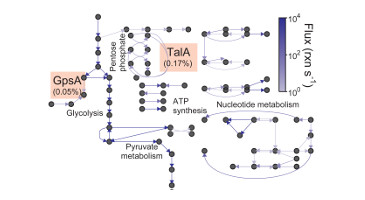
TR&D 1 initially started out as a project to develop tools for scalably aggregating data for modeling, organizing this data for model design, and systematically designing models from this data. This resulted in a number of tools that were developed with our collaborators. Some of these include:
1. Datanator:
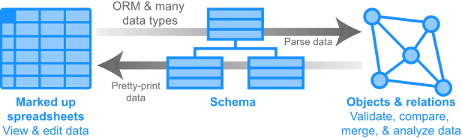
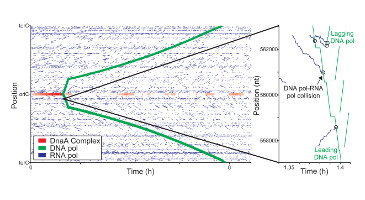
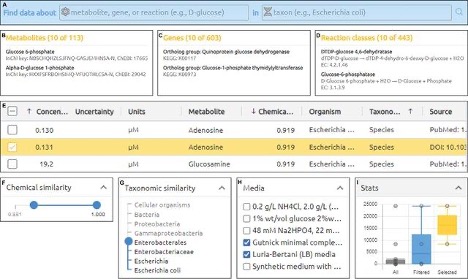
Datanator is an integrated database of molecular data designed to facilitate integrative analysis of multiple cellular subsystems. To help investigators overcome the lack of complete data about every molecule and reaction in every organism, Datanator includes tools for building “clouds” of data that include data about chemically similar molecules and reactions in phylogenetically similar organisms.
GitHub site: https://github.com/KarrLab/datanator
2. ObjTables:
ObjTables is a community initiative to make supplementary spreadsheets to journal articles – one of the most common methods for sharing scientific data – more reusable. The long-term goal is to create an ecosystem of reusable data for comparative and integrative research.
In year 2.5, TR&D 1 pivoted to developing the biosimulations portal.
GitHub site: https://github.com/KarrLab/obj_tables
BioSimulations is a free platform for sharing and re-using biomodels, simulations, simulation results, and visualizations of simulation results. BioSimulations supports a wide range of frameworks (e.g., logical, Flux-Balance Analysis (FBA), continuous kinetic, discrete kinetic), simulation algorithms (e.g., FBA, SSA), model formats (e.g., SBML), and tools (e.g., COBRApy, COPASI, tellurium).
Biosimulations may be considered a reproducibility portal for biomedical models. Our current work (August 2022) involves curating over 100 models so that a researcher may search for a given paper on biosimulations.org and reproduce the figures found in that paper.
Moving Forward: With reproducibility of biomedical models technically resolved, we are now switching our efforts to a related topic but even more important, model credibility. Model credibility (or lack of) became more obvious during the covid pandemic when many models of covid spread, and infection came under intense scrutiny. Model credibility is related to many factors, including validation, verification, sources of data, biophysical appropriateness, etc. Each of these factors, as well as others, contributes to an overall measure of credibility or trust in a model.
Although technical resolved, many published models remain non-reproducible and we discovered it is extremely hard to persuade authors and journals to ensure that their models are reproducible. Instead, a measure of a model’s credibility may be of more interest to researchers.
Software
ObjTables
https://youtu.be/OJmqtJey4zg ObjTables is a community initiative to make supplementary spreadsheets to journal articles - one of the most common methods for sharing scientific data - more reusable. The long-term goal is to create [...]
Datanator
Danator is an integrated database of molecular data designed to facilitate integrative analysis of multiple cellular subsystems. To help investigators overcome the lack of complete data about every molecule and reaction in every organism, [...]
Kinetic Datanator
Kinetic Datanator is a tool for discovering the data needed to build, calibrate, and validate whole-cell models. Kinetic Datanator is composed of an integrated database of experimental data for whole-cell modeling and tools for [...]
Random WC model generator
The Random Whole-cell model generator is a tool for generating WC models that represent user-specified numbers of genes, RNA, proteins, and reactions of an archetypal bacterium. The models generated by the model generator represent [...]
WC-KB
Whole-cell KB (WC-KB) is a data model for describing the experimental data needed to build, calibrate, and validate a whole-cell model. WC-KB includes a command line program and a Python API. Whole-cell KB [...]
WC-KB-Gen
WC-KB-Gen is a Python framework for programmatically constructing knowledge bases for whole-cell models. WC-KB-Gen helps modelers retrieve data from external sources, organize this data, and record the provenance of this data. In turn, WC-KB-Gen [...]
WC-Lang
WC-Lang is a data model and a file format for describing composite, multi-algorithmic whole-cell models. WC-Lang includes a command line interface and a Python API. WC-Lang
WC-Model-Gen
WC-Model-Gen is a Python framework for programmatically constructing whole-cell models from large datasets described with WC-KB. WC-Model-Gen helps modelers retrieve data from a WC-KB and use the data to scalably design species and reactions. [...]
WC-Rules
WC-Rules is a formalism for describing composite, mixed-grained, multi-algorithmic WC models. WC-Rules provides modelers a high-level, biologically-intuitive language for describing models in terms of patterns of metabolite, DNA, RNA, and protein species, and rules [...]
Collaborative projects
Multiscale, systems modeling of macrophage infection
Markus Covert Stanford University Stanford, CA, USA
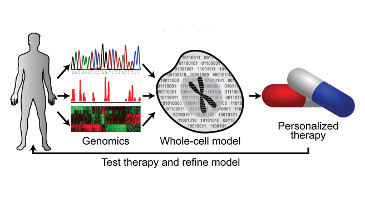
Toward whole-cell models for precision medicine
Marc Birtwistle and Jianlong Wang Icahn school of Medicine at Mount Sinai New York, NY, USA
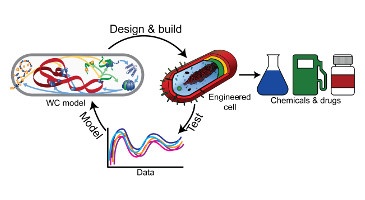
MiniCell – A model-driven approach to minimal cell engineering for medicine
Luis Serrano and Maria Lluch-Senar Center for Regulatory Genomics Barcelona, Spain Alain Blanchard and Carole Lartigue National Institute for Agricultural Research Bordeaux, France Jörg Stülke University of Göttingen Gottingen, Germany
Service projects
Alzheimer modeling
Jean-Marie Bouteiller Assistant Professor Department of Biomedical Engineering University of Southern California Los Angeles, CA, USA
CompuCell3d multiscale modeling
Drs. Maciek Swat and James Glazier The Biocomplexity Institute Indiana University Bloomington, IN, USA
PRiME: Program for Research on Immune Modeling and Experimentation
Stuart Sealfon Professor Neurology, Neurobiology, and Pharmacology & Systems Therapeutics Icahn School of Medicine at Mount Sinai New York, NY, USA
Simtk
Dr. Joy Ku Stanford University Stanford, CA, USA
Systems Biology Center New York
Ravi Iyengar Professor, Department of Pharmacology and Systems Therapeutics Icahn School of Medicine at Mount Sinai New York, NY, USA