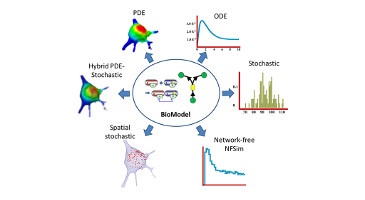
Models represent our knowledge, observations and hypotheses in a testable digital framework. Because models are digital, it should be easy to reuse models and reproduce simulation results. However, many dynamic biochemical models are not reusable and many simulation results are not reproducible, including models and simulation results reported in standard formats such as the Systems Biology Markup Language (SBML) and the Simulation Experiment Description Markup Language (SED-ML). This irreproducibility limits the impact of modeling by inhibiting researchers from reusing models and simulation results for additional studies and combining models of individual biological processes into meta-models of entire biological systems.
Currently, models are hard to reuse and simulations are hard to reproduce because (a) few researchers report the metadata needed to reproduce simulations, (b) there are many incompatible simulators, (c) there is no simulation results repository, (d) there is no standard for reducing simulation results, (e) there is no standard for describing results visualizations, and (f) there are inadequate tools for visualizing simulation results.
To address these problems, we will develop novel tools and public servers for (a) using existing simulators to reproducibly simulate a wide range of models and (b) storing and (c) visualizing simulation results:
- We will build a database for storing models, simulation experiments, their results, and their metadata which will mint DOIs and support queries over simulation results. The system will help researchers share and retrieve simulation results and apply big data analytics to simulation results. In turn, the system will help researchers reuse simulation experiments and reproduce simulation results.
- We will build a simulation system which provides a common interface to multiple simulators that each support individual simulation algorithms and modeling domains. This will make it easy for researchers to reuse models and reproduce simulations without having to install domain-specific simulators.
- We will build a web-based system for using the simulation system and simulation results database to interactively simulate and visualize models in a browser. This will enable researchers to retrieve deposited simulation results, request new simulations, and visually analyze simulation results.
To ensure our tools advance biomodeling, we will develop our tools in conjunction with several CPs and SPs which will provide model repositories and journals web-based tools for interactively simulating and visualizing reported models. These CPs will push us to develop user-friendly tools, and we will pull the CPs to require model authors to annotate their simulation experiments so they are reproducible.
To help researchers use our software, we will work with TR&Ds 1 and 2 to combine our software into a reproducible modeling workflow. We will also extensively document our software and distribute it open-source. In addition, as part of the Training and Dissemination Core, we will develop tutorials and organize workshops.
Software Tools and Standards
Formalized OMEX metadata specification
As a common language to represent annotations for models, we have developed, in close collaboration with the COMBINE community, the OMEX metadata specification. This standard provides not only the specification of sharable, RDF formatted [...]
Enhanced SED-ML libraries and specification
The Simulation Experiment Description Markup Language (SED-ML) is a standardized, community-based format that explicitly describes simulation processes for biochemical models, ensuring that researchers who are interested in a published model can efficiently reproduce and [...]
BioSimulations
BioSimulations is a free platform for sharing and re-using biomodels, simulations, simulation results, and visualizations of simulation results. BioSimulations supports a wide range of frameworks (e.g., logical, Flux-Balance Analysis (FBA), continuous kinetic, discrete kinetic), [...]
runBioSimulations
runBioSimulations is a free tool for running a wide range of biological simulations. Through the BioSimulators registry of biosimulation tools, runBioSimulations supports a broad range of modeling frameworks (e.g., logical, kinetic), simulation algorithms (e.g., FBA, SSA), [...]
BioSimulators
BioSimulators is a free registry of biosimulation tools. The registry includes tools for a broad range of frameworks (e.g., logical, kinetic), simulation algorithms (e.g., FBA, SSA), and model formats (e.g., BNGL, CellML, NeuroML/LEMS, SBML, Smoldyn). [...]
libRoadRunner
libRoadRunner is a high-performance and portable simulation engine for systems and synthetic biology. It can run on many platforms including Windows, Mac OS, and Linux. libRoadRunner is a major rewrite of the original C# [...]
VCell
VCell (Virtual Cell) is a comprehensive platform for modeling cell biological systems that are built on a central database and disseminated as a web application. VCell permits the construction of models, application of numerical [...]
WholeCellSimDB
WholeCellSimDB.org is a database of whole-cell model simulations designed to make it easy for researchers to explore and analyze whole-cell model predictions including predicted: Metabolite concentrations, DNA, RNA and protein expression, DNA-bound protein positions, [...]
WholeCellViz
WholeCellViz.org is a web-based software program for visually analyzing whole-cell simulations. WholeCellViz enables users to interactively analyze many aspects of cell physiology including: Cell mass, volume, and shape, Metabolite, RNA, and protein copy numbers, [...]
Collaborative projects
Alzheimer modeling
Jean-Marie Bouteiller Assistant Professor Department of Biomedical Engineering University of Southern California Los Angeles, CA, USA
BioModels model repository
Henning Hermjakob The European Bioinformatics Institute Cambridge, UK
CompuCell3d multiscale modeling
Drs. Maciek Swat and James Glazier The Biocomplexity Institute Indiana University Bloomington, IN, USA
JWS Online modeling environment
Jacky Snoep Stellenbosch University Stellenbosch, South Africa
PRiME: Program for Research on Immune Modeling and Experimentation
Stuart Sealfon Professor Neurology, Neurobiology, and Pharmacology & Systems Therapeutics Icahn School of Medicine at Mount Sinai New York, NY, USA
Simtk
Dr. Joy Ku Stanford University Stanford, CA, USA
Systems Biology Center New York
Ravi Iyengar Professor, Department of Pharmacology and Systems Therapeutics Icahn School of Medicine at Mount Sinai New York, NY, USA
Virtual Cell modeling environment
Leslie Loew Professor University of Connecticut Health Center Farmington, CT, USA
Service projects
PRiME: Program for Research on Immune Modeling and Experimentation
Stuart Sealfon Professor Neurology, Neurobiology, and Pharmacology & Systems Therapeutics Icahn School of Medicine at Mount Sinai New York, NY, USA
Simtk
Dr. Joy Ku Stanford University Stanford, CA, USA
Systems Biology Center New York
Ravi Iyengar Professor, Department of Pharmacology and Systems Therapeutics Icahn School of Medicine at Mount Sinai New York, NY, USA