 Recently published in Current Opinions in Systems Biology:
Recently published in Current Opinions in Systems Biology:
Discrete, logic-based models are increasingly used to describe biological mechanisms. Initially introduced to study gene regulation, these models evolved to cover various molecular mechanisms, such as signaling, transcription factor cooperativity, and even metabolic processes. The abstract nature and amenability of discrete models to robust mathematical analyses make them appropriate for addressing a wide range of complex biological problems.
Recent technological breakthroughs have generated a wealth of high throughput data. Novel, literature-based representations of biological processes and emerging algorithms offer new opportunities for model construction. Here, the authors review up-to-date efforts to address challenging biological questions by incorporating omic data into logic-based models, and discuss critical difficulties in constructing and analyzing integrative, large-scale, logic-based models of biological mechanisms.
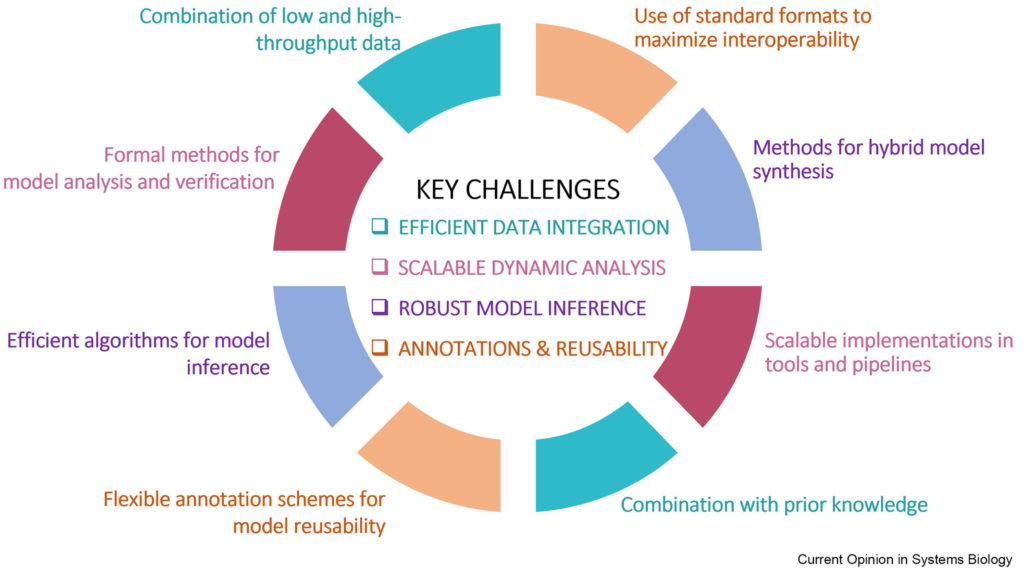
Figure: Different data types and sources and their uses in the inference and analysis of logic-based models.

