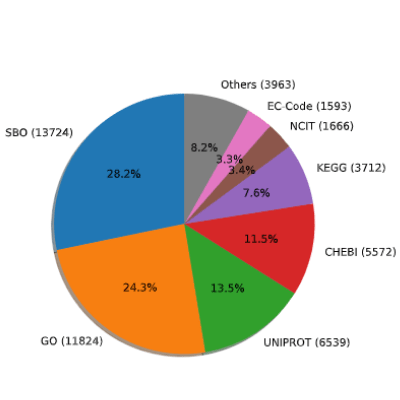
Distribution of knowledge resources of the four element types, obtained from 1,000 BioModels.Only seven most frequently used knowledge resources were specified. Annotations were collected either by thesbotermattribute or thegetAnnotationString()method. In annotation strings, those with eitherbqbiol:isorbqbiol:isVersionOftag were selected. SBO and GO consisted more than 50% of all annotations found. In thefollowing analyses, we used the five most common ontologies, i.e., SBO, GO, UNIPROT, CHEBI, and KEGG.
The interests in repurposing and reusing systems biology models have been growing in recent years. Semantic annotations play an important role for this, as they provide crucial information on the meanings and functions of models. However, there are a limited number of tools that evaluate the existence or quality of such annotations. In this paper, the authors introduce SBMate, a python package that would serve as a framework for evaluating the quality of annotations in systems biology models. Three default metrics are provided: coverage, consistency, and specificity. Coverage checks whether annotations exist in a model. Consistency tests if the annotations are appropriate for the given model element. Finally, specificity represents how detailed the annotations are. The research analyzed 1,000 curated models from the BioModels repository using the three metrics and discussed the results. Additional metrics can be easily added to extend the current version of SBMate.
Preprints and early-stage research may not have been peer-reviewed yet.

